diff --git a/source/Tutorials/DP-GEN/DP-GEN_handson.md b/source/Tutorials/DP-GEN/DP-GEN_handson.md
new file mode 100644
index 0000000..dee82b9
--- /dev/null
+++ b/source/Tutorials/DP-GEN/DP-GEN_handson.md
@@ -0,0 +1,433 @@
+# Hands-on tutorial for DP-GEN (v0.10.3)
+
+Writer: Wenshuo Liang
+
+Verifier: Yibo Wang
+
+## General Introduction
+Deep Potential GENerator (DP-GEN) is a package that implements a concurrent learning scheme to generate reliable DP models. Typically, the DP-GEN workflow contains three processes: init, run, and autotest.
+
+- init: generate the initial training dataset by first-principle calculations.
+- run: the main process of DP-GEN, in which the training dataset is enriched and the quality of the DP models is improved automatically.
+- autotest: calculate a simple set of properties and/or perform tests for comparison with DFT and/or empirical interatomic potentials.
+
+This is a practical tutorial that aims to help you quickly get command of dpgen run interface.
+
+## Input files
+In this tutorial, we take a gas-phase methane molecule as an example. We have prepared input files in dpgen_example/run
+
+Now download the dpgen_example and uncompress it:
+
+```sh
+wget https://dp-public.oss-cn-beijing.aliyuncs.com/community/dpgen_example.tar.xz
+tar xvf dpgen_example.tar.xz
+```
+Now go into the dpgen_example/run.
+```sh
+$ cd dpgen_example/run
+$ ls
+INCAR_methane machine.json param.json POTCAR_C POTCAR_H
+```
+
+- param.json is the settings for DP-GEN for the current task. It will expalin later.
+- machine.json is a task dispatcher where the machine environment and resource requirements are set.[We have expalined here](www.baidu.com)
+- INCAR* and POTCAR* are the input file for the VASP package. All first-principle calculations share the same parameters as the one you set in param.json.
+
+## Run process
+
+We can run DP-GEN easily by:
+```sh
+$ dpgen run param.json machine.json
+```
+
+The run process contains a series of successive iterations. Each iteration is composed of three steps:
+
+* `exploration`
+* `labeling`
+* `training`
+
+Accordingly, there are three sub-folders in each iteration:
+
+* `00.train`
+* `01.model_devi`
+* `02.fp`
+
+
+### param.json
+
+ We provide an example of param.json.
+
+```json
+{
+ "type_map": ["H","C"],
+ "mass_map": [1,12],
+ "init_data_prefix": "../",
+ "init_data_sys": ["init/CH4.POSCAR.01x01x01/02.md/sys-0004-0001/deepmd"],
+ "sys_configs_prefix": "../",
+ "sys_configs": [
+ ["init/CH4.POSCAR.01x01x01/01.scale_pert/sys-0004-0001/scale/00000/POSCAR"],
+ ["init/CH4.POSCAR.01x01x01/01.scale_pert/sys-0004-0001/scale/00001/POSCAR"]
+ ],
+ "_comment": " that's all ",
+ "numb_models": 4,
+ "default_training_param": {
+ "model": {
+ "type_map": ["H","C"],
+ "descriptor": {
+ "type": "se_a",
+ "sel": [16,4],
+ "rcut_smth": 0.5,
+ "rcut": 5.0,
+ "neuron": [120,120,120],
+ "resnet_dt": true,
+ "axis_neuron": 12,
+ "seed": 1
+ },
+ "fitting_net": {
+ "neuron": [25,50,100],
+ "resnet_dt": false,
+ "seed": 1
+ }
+ },
+ "learning_rate": {
+ "type": "exp",
+ "start_lr": 0.001,
+ "decay_steps": 5000
+ },
+ "loss": {
+ "start_pref_e": 0.02,
+ "limit_pref_e": 2,
+ "start_pref_f": 1000,
+ "limit_pref_f": 1,
+ "start_pref_v": 0.0,
+ "limit_pref_v": 0.0
+ },
+ "training": {
+ "stop_batch": 400000,
+ "disp_file": "lcurve.out",
+ "disp_freq": 1000,
+ "numb_test": 4,
+ "save_freq": 1000,
+ "save_ckpt": "model.ckpt",
+ "disp_training": true,
+ "time_training": true,
+ "profiling": false,
+ "profiling_file": "timeline.json",
+ "_comment": "that's all"
+ }
+ },
+ "model_devi_dt": 0.002,
+ "model_devi_skip": 0,
+ "model_devi_f_trust_lo": 0.05,
+ "model_devi_f_trust_hi": 0.15,
+ "model_devi_e_trust_lo": 10000000000.0,
+ "model_devi_e_trust_hi": 10000000000.0,
+ "model_devi_clean_traj": true,
+ "model_devi_jobs": [
+ {"sys_idx": [0],"temps": [100],"press": [1.0],"trj_freq": 10,"nsteps": 300,"ensemble": "nvt","_idx": "00"},
+ {"sys_idx": [1],"temps": [100],"press": [1.0],"trj_freq": 10,"nsteps": 3000,"ensemble": "nvt","_idx": "01"}
+ ],
+ "fp_style": "vasp",
+ "shuffle_poscar": false,
+ "fp_task_max": 20,
+ "fp_task_min": 5,
+ "fp_pp_path": "./",
+ "fp_pp_files": ["POTCAR_H","POTCAR_C"],
+ "fp_incar": "./INCAR_methane"
+}
+```
+
+The following is a detailed description of the keywords.
+
+#### Basics keywords (Line 2-3):
+
+| Key | Type | Description |
+|-------------|-----------------|-------------------------|
+| "type_map" | List of string | Atom types |
+| "mass_map" | List of float | Standard atom weights. |
+
+
+#### Data-related keywords (Line 4-10):
+| Key | Type | Description |
+|-----------------------|-----------------|-------------------------------------------------------------------------------------------------------------|
+| "init_data_prefix" | String | Prefix of initial data directories |
+| "init_data_sys" | List of string | Directories of initial data. You may use either absolute or relative path here. |
+| "sys_configs_prefix" | String | Prefix of sys_configs |
+| "sys_configs" | List of string | Containing directories of structures to be explored in iterations. Wildcard characters are supported here. |
+
+#### Training-related keywords (Line 12-58):
+| Key | Type | Description |
+|---------------------------|----------|----------------------------------------------|
+| "numb_models" | Integer | Number of models to be trained in 00.train. |
+| "default_training_param" | Dict | Training parameters for deepmd-kit. |
+
+#### Exploration-related keywords (Line 59-69):
+| Key | Type | Description |
+|--------------------------|-------------------------|-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|
+| "model_devi_dt" | Float | Timestep for MD |
+| "model_devi_skip" | Integer | Number of structures skipped for fp in each MD |
+| "model_devi_f_trust_lo" | Float | Lower bound of forces for the selection. If List, should be set for each index in sys_configs, respectively. |
+| "model_devi_f_trust_hi" | Integer | Upper bound of forces for the selection. If List, should be set for each index in sys_configs, respectively. |
+| "model_devi_f_trust_hi" | Float or List of float | Lower bound of virial for the selection. If List, should be set for each index in sys_configs, respectively. Should be used with DeePMD-kit v2.x. |
+| "model_devi_f_trust_hi" | Float or List of float | Upper bound of virial for the selection. If List, should be set for each index in sys_configs, respectively. Should be used with DeePMD-kit v2.x. |
+| "model_devi_clean_traj" | Boolean or Int | If type of model_devi_clean_traj is boolean type then it denote whether to clean traj folders in MD since they are too large. If it is Int type, then the most recent n iterations of traj folders will be retained, others will be removed. |
+| "model_devi_jobs" | List of Dict | Settings for exploration in 01.model_devi. Each dict in the list corresponds to one iteration. The index of model_devi_jobs exactly accord with index of iterations |
+
+#### Labeling-related parameters (Line 70-76):
+| Key | Type | Description |
+|-------------------|-----------------|--------------------------------------------------------------------------------------------------------------------------|
+| "fp_style" | String | Software for First Principles. Options include “vasp”, “pwscf”, “siesta” and “gaussian” up to now. |
+| "shuffle_poscar" | Boolean | |
+| "fp_task_max" | Integer | Maximum of structures to be calculated in 02.fp of each iteration. |
+| "fp_task_min" | Integer | Minimum of structures to calculate in 02.fp of each iteration. |
+| "fp_pp_path" | String | Directory of psuedo-potential file to be used for 02.fp exists. |
+| "fp_pp_files" | List of string | Psuedo-potential file to be used for 02.fp. Note that the order of elements should correspond to the order in type_map. |
+| "fp_incar" | String | Input file for VASP. INCAR must specify KSPACING and KGAMMA. |
+
+## Output files
+
+In dpgen_example/run, we can find that a folder and two files are generated automatically.
+```sh
+$ ls
+dpgen.log INCAR_methane iter.000000 machine.json param.json record.dpgen
+```
+
+- `iter.000000` contains the main results that DP-GEN generates in the first iteration.
+- `record.dpgen` records the current stage of the run process.
+- `dpgen.log` includes time and iteration information.
+ When the first iteration is completed, the folder structure of `iter.000000` is like this:
+
+```sh
+$ tree iter.000000/ -L 1
+./iter.000000/
+├── 00.train
+├── 01.model_devi
+└── 02.fp
+```
+### 00.train
+First, we check the folder `iter.000000`/ `00.train`.
+```sh
+$ tree iter.000000/00.train -L 1
+./iter.000000/00.train/
+├── 000
+├── 001
+├── 002
+├── 003
+├── data.init -> /root/dpgen_example
+├── data.iters
+├── graph.000.pb -> 000/frozen_model.pb
+├── graph.001.pb -> 001/frozen_model.pb
+├── graph.002.pb -> 002/frozen_model.pb
+└── graph.003.pb -> 003/frozen_model.pb
+```
+
+- Folder 00x contains the input and output files of the DeePMD-kit, in which a model is trained.
+- graph.00x.pb , linked to 00x/frozen.pb, is the model DeePMD-kit generates. The only difference between these models is the random seed for neural network initialization.
+We may randomly select one of them, like 000.
+```sh
+$ tree iter.000000/00.train/000 -L 1
+./iter.000000/00.train/000
+├── checkpoint
+├── frozen_model.pb
+├── input.json
+├── lcurve.out
+├── model.ckpt-400000.data-00000-of-00001
+├── model.ckpt-400000.index
+├── model.ckpt-400000.meta
+├── model.ckpt.data-00000-of-00001
+├── model.ckpt.index
+├── model.ckpt.meta
+└── train.log
+```
+
+- `input.json` is the settings for deepmd-kit for current task.
+- `checkpoint` is used for restart traning.
+- `model.ckpt*` are model related files.
+- `frozen_model.pb` is the frozen model.
+- `lcurve.out` records the training accuracy of energies and forces.
+- `train.log` includes version, data, hardware information, time, etc.
+
+### 01.model_devi
+Then, we check the folder iter.000000/ 01.model_devi.
+```sh
+$ tree iter.000000/01.model_devi -L 1
+./iter.000000/01.model_devi/
+├── confs
+├── graph.000.pb -> /root/dpgen_example/run/iter.000000/00.train/graph.000.pb
+├── graph.001.pb -> /root/dpgen_example/run/iter.000000/00.train/graph.001.pb
+├── graph.002.pb -> /root/dpgen_example/run/iter.000000/00.train/graph.002.pb
+├── graph.003.pb -> /root/dpgen_example/run/iter.000000/00.train/graph.003.pb
+├── task.000.000000
+├── task.000.000001
+├── task.000.000002
+├── task.000.000003
+├── task.000.000004
+├── task.000.000005
+├── task.000.000006
+├── task.000.000007
+├── task.000.000008
+└── task.000.000009
+```
+
+- Folder confs contains the initial configurations for LAMMPS MD converted from POSCAR you set in "sys_configs" of param.json.
+
+- Folder task.000.00000x contains the input and output files of the LAMMPS. We may randomly select one of them, like task.000.000001.
+```sh
+$ tree iter.000000/01.model_devi/task.000.000001
+./iter.000000/01.model_devi/task.000.000001
+├── conf.lmp -> ../confs/000.0001.lmp
+├── input.lammps
+├── log.lammps
+├── model_devi.log
+└── model_devi.out
+```
+
+- `conf.lmp`, linked to `000.0001.lmp` in folder confs, serves as the initial configuration of MD.
+- `input.lammps` is the input file for LAMMPS.
+- `model_devi.out` records the model deviation of concerned labels, energy and force, in MD. It serves as the criterion for selecting which structures and doing first-principle calculations.
+
+By head `model_devi.out`, you will see:
+```
+$ head -n 5 ./iter.000000/01.model_devi/task.000.000001/model_devi.out
+ # step max_devi_v min_devi_v avg_devi_v max_devi_f min_devi_f avg_devi_f
+ 0 1.438427e-04 5.689551e-05 1.083383e-04 8.835352e-04 5.806717e-04 7.098761e-04
+10 3.887636e-03 9.377374e-04 2.577191e-03 2.880724e-02 1.329747e-02 1.895448e-02
+20 7.723417e-04 2.276932e-04 4.340100e-04 3.151907e-03 2.430687e-03 2.727186e-03
+30 4.962806e-03 4.943687e-04 2.925484e-03 5.866077e-02 1.719157e-02 3.011857e-02
+```
+Now we'll concentrate on `max_devi_f`.
+Recall that we've set `"trj_freq"` as 10, so every 10 steps the structures are saved. Whether to select the structure depends on its `"max_devi_f"`. If it falls between `"model_devi_f_trust_lo"` (0.05) and `"model_devi_f_trust_hi"` (0.15), DP-GEN will treat the structure as a candidate. Here, only the 30th structure will be selected, whose `"max_devi_f"` is 5.866077e e-02.
+
+### 02.fp
+Finally, we check the folder iter.000000/ 02.fp.
+```
+$ tree iter.000000/02.fp -L 1
+./iter.000000/02.fp
+├── data.000
+├── task.000.000000
+├── task.000.000001
+├── task.000.000002
+├── task.000.000003
+├── task.000.000004
+├── task.000.000005
+├── task.000.000006
+├── task.000.000007
+├── task.000.000008
+├── task.000.000009
+├── task.000.000010
+├── task.000.000011
+├── candidate.shuffled.000.out
+├── POTCAR.000
+├── rest_accurate.shuffled.000.out
+└── rest_failed.shuffled.000.out
+```
+
+- `POTCAR` is the input file for VASP generated according to `"fp_pp_files"` of param.json.
+- `candidate.shuffle.000.out` records which structures will be selected from last step 01.model_devi. There are always far more candidates than the maximum you expect to calculate at one time. In this condition, DP-GEN will randomly choose up to `"fp_task_max"` structures and form the folder task.*.
+- `rest_accurate.shuffle.000.out` records the other structures where our model is accurate ("max_devi_f" is less than `"model_devi_f_trust_lo"`, no need to calculate any more),
+- `rest_failed.shuffled.000.out` records the other structures where our model is too inaccurate (lager than `"model_devi_f_trust_hi"`, there may be some error).
+- `data.000`: After first-principle calculations, DP-GEN will collect these data and change them into the format DeePMD-kit needs. In the next iteration's `00.train`, these data will be trained together as well as initial data.
+
+By cat candidate.shuffled.000.out \| grep task.000.000001, you will see:
+
+```sh
+$ cat ./iter.000000/02.fp/candidate.shuffled.000.out | grep task.000.000001
+iter.000000/01.model_devi/task.000.000001 190
+iter.000000/01.model_devi/task.000.000001 130
+iter.000000/01.model_devi/task.000.000001 120
+iter.000000/01.model_devi/task.000.000001 150
+iter.000000/01.model_devi/task.000.000001 280
+iter.000000/01.model_devi/task.000.000001 110
+iter.000000/01.model_devi/task.000.000001 30
+iter.000000/01.model_devi/task.000.000001 230
+```
+
+The `task.000.000001` 30 is exactly what we have just found in `01.model_devi` satisfying the criterion to be calculated again.
+After the first iteration, we check the contents of dpgen.log and record.dpgen.
+
+```sh
+$ cat dpgen.log
+2022-03-07 22:12:45,447 - INFO : start running
+2022-03-07 22:12:45,447 - INFO : =============================iter.000000==============================
+2022-03-07 22:12:45,447 - INFO : -------------------------iter.000000 task 00--------------------------
+2022-03-07 22:12:45,451 - INFO : -------------------------iter.000000 task 01--------------------------
+2022-03-08 00:53:00,179 - INFO : -------------------------iter.000000 task 02--------------------------
+2022-03-08 00:53:00,179 - INFO : -------------------------iter.000000 task 03--------------------------
+2022-03-08 00:53:00,187 - INFO : -------------------------iter.000000 task 04--------------------------
+2022-03-08 00:57:04,113 - INFO : -------------------------iter.000000 task 05--------------------------
+2022-03-08 00:57:04,113 - INFO : -------------------------iter.000000 task 06--------------------------
+2022-03-08 00:57:04,123 - INFO : system 000 candidate : 12 in 310 3.87 %
+2022-03-08 00:57:04,125 - INFO : system 000 failed : 0 in 310 0.00 %
+2022-03-08 00:57:04,125 - INFO : system 000 accurate : 298 in 310 96.13 %
+2022-03-08 00:57:04,126 - INFO : system 000 accurate_ratio: 0.9613 thresholds: 1.0000 and 1.0000 eff. task min and max -1 20 number of fp tasks: 12
+2022-03-08 00:57:04,154 - INFO : -------------------------iter.000000 task 07--------------------------
+2022-03-08 01:02:07,925 - INFO : -------------------------iter.000000 task 08--------------------------
+2022-03-08 01:02:07,926 - INFO : failed tasks: 0 in 12 0.00 %
+2022-03-08 01:02:07,949 - INFO : failed frame: 0 in 12 0.00 %
+```
+
+It can be found that 310 structures are generated in iter.000000, in which 12 structures are collected for first-principle calculations.
+```sh
+$ cat record.dpgen
+0 0
+0 1
+0 2
+0 3
+0 4
+0 5
+0 6
+0 7
+0 8
+```
+
+Each line contains two numbers: the first is the index of iteration, and the second, ranging from 0 to 9, records which stage in each iteration is currently running.
+
+| Index of iterations | "Stage in eachiteration " | Process |
+|----------------------|-----------------------------|------------------|
+| 0 | 0 | make_train |
+| 0 | 1 | run_train |
+| 0 | 2 | post_train |
+| 0 | 3 | make_model_devi |
+| 0 | 4 | run_model_devi |
+| 0 | 5 | post_model_devi |
+| 0 | 6 | make_fp |
+| 0 | 7 | run_fp |
+| 0 | 8 | post_fp |
+
+If the process of DP-GEN stops for some reason, DP-GEN will automatically recover the main process by record.dpgen. You may also change it manually for your purpose, such as removing the last iterations and recovering from one checkpoint.
+After all iterations, we check the structure of dpgen_example/run
+```sh
+$ tree ./ -L 2
+./
+├── dpgen.log
+├── INCAR_methane
+├── iter.000000
+│ ├── 00.train
+│ ├── 01.model_devi
+│ └── 02.fp
+├── iter.000001
+│ ├── 00.train
+│ ├── 01.model_devi
+│ └── 02.fp
+├── iter.000002
+│ └── 00.train
+├── machine.json
+├── param.json
+└── record.dpgen
+```
+
+and contents of `dpgen.log`.
+```sh
+$ cat cat dpgen.log | grep system
+2022-03-08 00:57:04,123 - INFO : system 000 candidate : 12 in 310 3.87 %
+2022-03-08 00:57:04,125 - INFO : system 000 failed : 0 in 310 0.00 %
+2022-03-08 00:57:04,125 - INFO : system 000 accurate : 298 in 310 96.13 %
+2022-03-08 00:57:04,126 - INFO : system 000 accurate_ratio: 0.9613 thresholds: 1.0000 and 1.0000 eff. task min and max -1 20 number of fp tasks: 12
+2022-03-08 03:47:00,718 - INFO : system 001 candidate : 0 in 3010 0.00 %
+2022-03-08 03:47:00,718 - INFO : system 001 failed : 0 in 3010 0.00 %
+2022-03-08 03:47:00,719 - INFO : system 001 accurate : 3010 in 3010 100.00 %
+2022-03-08 03:47:00,722 - INFO : system 001 accurate_ratio: 1.0000 thresholds: 1.0000 and 1.0000 eff. task min and max -1 0 number of fp tasks: 0
+```
+It can be found that 3010 structures are generated in `iter.000001`, in which no structure is collected for first-principle calculations. Therefore, the final models are not updated in iter.000002/00.train.
+
+
diff --git a/source/Tutorials/DP-GEN/index.rst b/source/Tutorials/DP-GEN/index.rst
index f5986cb..8cafa63 100644
--- a/source/Tutorials/DP-GEN/index.rst
+++ b/source/Tutorials/DP-GEN/index.rst
@@ -27,4 +27,6 @@ This tutorial tell you how to use DP-GEN,for detail information, you can check
.. toctree::
:maxdepth: 2
- :caption: Hands-on Tutorials
\ No newline at end of file
+ :caption: Hands-on Tutorials
+
+ learnDoc/DP-GEN_handson
\ No newline at end of file
diff --git a/source/Tutorials/DeePMD-kit/learnDoc/Handson-Tutorial(v2.0.3).md b/source/Tutorials/DeePMD-kit/learnDoc/Handson-Tutorial(v2.0.3).md
index 1d14594..2cd66e5 100644
--- a/source/Tutorials/DeePMD-kit/learnDoc/Handson-Tutorial(v2.0.3).md
+++ b/source/Tutorials/DeePMD-kit/learnDoc/Handson-Tutorial(v2.0.3).md
@@ -1,15 +1,30 @@
# Handson-Tutorial(v2.0.3)
-This tutorial will introduce you to the basic usage of the DeePMD-kit, taking a gas phase methane molecule as an example. Typically the DeePMD-kit workflow contains three parts: data preparation, training/freezing/compressing/testing, and molecular dynamics.
+This tutorial will introduce you to the basic usage of the DeePMD-kit, taking a gas phase methane molecule as an example. Typically the DeePMD-kit workflow contains three parts:
+
+1. Data preparation
+2. Training/Freezing/Compressing/Testing
+3. Molecular dynamics
The DP model is generated using the DeePMD-kit package (v2.0.3). The training data is converted into the format of DeePMD-kit using a tool named dpdata (v0.2.5). It needs to be noted that dpdata only works with Python 3.5 and later versions. The MD simulations are carried out using LAMMPS (29 Sep 2021) integrated with DeePMD-kit. Details of dpdata and DeePMD-kit installation and execution of can be found in [the DeepModeling official GitHub site](https://github.com/deepmodeling). OVITO is used for the visualization of the MD trajectory.
-The files needed for this tutorial are available [here](https://github.com/likefallwind/DPExample/raw/main/CH4.zip). The folder structure of this tutorial is like this:
+The files needed for this tutorial are available.
+```
+ $ wget https://dp-public.oss-cn-beijing.aliyuncs.com/community/CH4.tar
+ $ tar xvf CH4.tar
+```
+
+The folder structure of this tutorial is like this:
+ $ cd CH4
$ ls
00.data 01.train 02.lmp
-where the folder 00.data contains the data, the folder 01.train contains an example input script to train a model with DeePMD-kit, and the folder 02.lmp contains LAMMPS example script for molecular dynamics simulation.
+There are 3 folders here:
+
+1. The folder 00.data contains the data
+2. The folder 01.train contains an example input script to train a model with DeePMD-kit
+3. The folder 02.lmp contains LAMMPS example script for molecular dynamics simulation
## Data preparation
The training data of the DeePMD-kit contains the atom type, the simulation box, the atom coordinate, the atom force, the system energy, and the virial. A snapshot of a molecular system that has this information is called a frame. A system of data includes many frames that share the same number of atoms and atom types. For example, a molecular dynamics trajectory can be converted into a system of data, with each time step corresponding to a frame in the system.
@@ -20,7 +35,7 @@ We provide a convenient tool named dpdata for converting the data produced by VA
As an example, go to the data folder:
- $ cd data
+ $ cd 00.data
$ ls
OUTCAR
@@ -35,30 +50,47 @@ then execute the following commands:
data = dpdata.LabeledSystem('OUTCAR', fmt = 'vasp/outcar')
print('# the data contains %d frames' % len(data))
-On the screen, you can see that the OUTCAR file contains 200 frames of data. We randomly pick 40 frames as validation data and the rest as training data. The parameter set\_size specifies the set size. The parameter prec specifies the precision of the floating point number.
+On the screen, you can see that the OUTCAR file contains 200 frames of data. We randomly pick 40 frames as validation data and the rest as training data.
- index_validation = np.random.choice(200,size=40,replace=False)
- index_training = list(set(range(200))-set(index_validation))
+ index_validation = np.random.choice(200,size=40,replace=False) # random choose 40 index for validation_data
+ index_training = list(set(range(200))-set(index_validation)) # other indexes are training_data
data_training = data.sub_system(index_training)
data_validation = data.sub_system(index_validation)
- data_training.to_deepmd_npy('00.data/training_data')
- data_validation.to_deepmd_npy('00.data/validation_data')
+ data_training.to_deepmd_npy('training_data') # all training data put into directory:"training_data"
+ data_validation.to_deepmd_npy('validation_data') # all validation data put into directory:"validation_data"
print('# the training data contains %d frames' % len(data_training))
print('# the validation data contains %d frames' % len(data_validation))
-The commands import a system of data from the OUTCAR (with format vasp/outcar ), and then dump it into the compressed format (numpy compressed arrays). The data in DeePMD-kit format is stored in the folder 00.data..
+The commands import a system of data from the OUTCAR (with format vasp/outcar ), and then dump it into the compressed format (numpy compressed arrays). The data in DeePMD-kit format is stored in the folder 00.data. Lets have a look:
- $ ls 00.data/training_data
+```
+ $ ls
+ OUTCAR training_data validation_data
+```
+The directories "training_data" and "validation_data" have similar structure, so we just explain "training_data":
+
+ $ ls training_data
set.000 type.raw type_map.raw
- $ cat 00.data/training_data/type.raw
- H C
-Since all frames in the system have the same atom types and atom numbers, we only need to specify the type information once for the whole system
+1. set.000: is a directory, contains data in compressed format (numpy compressed arrays).
+2. type.raw: is a file, contains types of atoms(Represented in integer)
+3. type_map.raw: is a file, contains type name of atoms.
- $ cat 00.data/type_map.raw
+Lets have a look at `type.raw`:
+```
+ $ cat training_data/type.raw
0 0 0 0 1
+```
+This tells us there are 5 atoms in this example, 4 atoms represented by type "0", and 1 atom represented by type "1".
+Sometimes one needs to map the integer types to atom name. The mapping can be given by the file `type_map.raw`
-where atom H is given type 0, and atom C is given type 1.
+
+ $ cat training_data/type_map.raw
+ H C
+
+This tells us the type "0" is named by "H", and the type "1" is named by "C".
+
+More detailed doc about Data conversion can be found [here](https://docs.deepmodeling.org/projects/deepmd/en/master/data/data-conv.html)
## Training
### Prepare input script
@@ -94,7 +126,7 @@ In the model section, the parameters of embedding and fitting networks are speci
"_comment": "that's all"'
},
-The se\_e2\_a descriptor is used to train the DP model. The item neurons set the size of the embedding and fitting network to [10, 20, 40] and [100, 100, 100], respectively. The components in  to smoothly go to zero from 0.5 to 6 Å.
+The `se\_e2\_a` descriptor is used to train the DP model. The item neurons set the size of the embedding and fitting network to [10, 20, 40] and [100, 100, 100], respectively. The components in
to smoothly go to zero from 0.5 to 6 Å.
+The `se\_e2\_a` descriptor is used to train the DP model. The item neurons set the size of the embedding and fitting network to [10, 20, 40] and [100, 100, 100], respectively. The components in  to smoothly go to zero from 0.5 to 6 Å.
The following are the parameters that specify the learning rate and loss function.
@@ -116,7 +148,7 @@ The following are the parameters that specify the learning rate and loss functio
"_comment": "that's all"
},
-In the loss function, pref\_e increases from 0.02 to 1
to smoothly go to zero from 0.5 to 6 Å.
The following are the parameters that specify the learning rate and loss function.
@@ -116,7 +148,7 @@ The following are the parameters that specify the learning rate and loss functio
"_comment": "that's all"
},
-In the loss function, pref\_e increases from 0.02 to 1 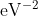 , and pref\_f decreases from 1000 to 1
, and pref\_f decreases from 1000 to 1 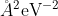 progressively, which means that the force term dominates at the beginning, while energy and virial terms become important at the end. This strategy is very effective and reduces the total training time. pref_v is set to 0
progressively, which means that the force term dominates at the beginning, while energy and virial terms become important at the end. This strategy is very effective and reduces the total training time. pref_v is set to 0 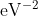 , indicating that no virial data are included in the training process. The starting learning rate, stop learning rate, and decay steps are set to 0.001, 3.51e-8, and 5000, respectively. The model is trained for
, indicating that no virial data are included in the training process. The starting learning rate, stop learning rate, and decay steps are set to 0.001, 3.51e-8, and 5000, respectively. The model is trained for 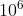 steps.
+In the loss function, `pref\_e` increases from 0.02 to 1
steps.
+In the loss function, `pref\_e` increases from 0.02 to 1 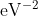 , and `pref\_f` decreases from 1000 to 1
, and `pref\_f` decreases from 1000 to 1 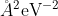 progressively, which means that the force term dominates at the beginning, while energy and virial terms become important at the end. This strategy is very effective and reduces the total training time. `pref_v` is set to 0
progressively, which means that the force term dominates at the beginning, while energy and virial terms become important at the end. This strategy is very effective and reduces the total training time. `pref_v` is set to 0 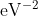 , indicating that no virial data are included in the training process. The starting learning rate, stop learning rate, and decay steps are set to 0.001, 3.51e-8, and 5000, respectively. The model is trained for
, indicating that no virial data are included in the training process. The starting learning rate, stop learning rate, and decay steps are set to 0.001, 3.51e-8, and 5000, respectively. The model is trained for 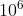 steps.
The training parameters are given in the following
@@ -176,7 +208,8 @@ If everything works fine, you will see, on the screen, information printed every
DEEPMD INFO batch 10000 training time 6.41 s, testing time 0.01 s
DEEPMD INFO saved checkpoint model.ckpt
-They present the training and testing time counts. At the end of the 10000th batch, the model is saved in Tensorflow's checkpoint file model.ckpt. At the same time, the training and testing errors are presented in file lcurve.out.
+They present the training and testing time counts. At the end of the 10000th batch, the model is saved in Tensorflow's checkpoint file `model.ckpt`. At the same time, the training and testing errors are presented in file `lcurve.out`.
+The file contains 8 columns, form left to right, are the training step, the validation loss, training loss, root mean square (RMS) validation error of energy, RMS training error of energy, RMS validation error of force, RMS training error of force and the learning rate. The RMS error (RMSE) of the energy is normalized by number of atoms in the system.
$ head -n 2 lcurve.out
#step rmse_val rmse_trn rmse_e_val rmse_e_trn rmse_f_val rmse_f_trn lr
@@ -190,11 +223,29 @@ and
Volumes 4, 5 and 6, 7 present energy and force training and testing errors, respectively. It is demonstrated that after 140,000 steps of training, the energy testing error is less than 1 meV and the force testing error is around 120 meV/Å. It is also observed that the force testing error is systematically (but slightly) larger than the training error, which implies a slight over-fitting to the rather small dataset.
+One can visualize this file by a simple Python script:
+
+```py
+import numpy as np
+import matplotlib.pyplot as plt
+
+data = np.genfromtxt("lcurve.out", names=True)
+for name in data.dtype.names[1:-1]:
+ plt.plot(data['step'], data[name], label=name)
+plt.legend()
+plt.xlabel('Step')
+plt.ylabel('Loss')
+plt.xscale('symlog')
+plt.yscale('log')
+plt.grid()
+plt.show()
+```
+
When the training process is stopped abnormally, we can restart the training from the provided checkpoint by simply running
$ dp train --restart model.ckpt input.json
-In the lcurve.out, you can see the training and testing errors, like
+In the `lcurve.out`, you can see the training and testing errors, like
538000 3.12e-01 2.16e-01 6.84e-04 7.52e-04 1.38e-01 9.52e-02 4.1e-06
538000 3.12e-01 2.16e-01 6.84e-04 7.52e-04 1.38e-01 9.52e-02 4.1e-06
@@ -203,7 +254,7 @@ In the lcurve.out, you can see the training and testing errors, like
530000 2.89e-01 2.15e-01 6.36e-04 5.18e-04 1.25e-01 9.31e-02 4.4e-06
531000 3.46e-01 3.26e-01 4.62e-04 6.73e-04 1.49e-01 1.41e-01 4.4e-06
-Note that input.json needs to be consistent with the previous one.
+Note that `input.json` needs to be consistent with the previous one.
### Freeze and Compress a model
At the end of the training, the model parameters saved in TensorFlow's checkpoint file should be frozen as a model file that is usually ended with extension .pb. Simply execute
@@ -212,8 +263,8 @@ At the end of the training, the model parameters saved in TensorFlow's checkpoin
DEEPMD INFO Restoring parameters from ./model.ckpt-1000000
DEEPMD INFO 1264 ops in the final graph
-and it will output a model file named graph.pb in the current directory.
-The compressed DP model typically speed up DP-based calculations by an order of magnitude faster, and consume an order of magnitude less memory. The graph.pb can be compressed in the following way:
+and it will output a model file named `graph.pb` in the current directory.
+The compressed DP model typically speed up DP-based calculations by an order of magnitude faster, and consume an order of magnitude less memory. The `graph.pb` can be compressed in the following way:
$ dp compress -i graph.pb -o graph-compress.pb
DEEPMD INFO stage 1: compress the model
@@ -227,7 +278,7 @@ The compressed DP model typically speed up DP-based calculations by an order of
DEEPMD INFO Restoring parameters from model-compression/model.ckpt
DEEPMD INFO 840 ops in the final graph
-and it will output a model file named graph-compress.pb.
+and it will output a model file named `graph-compress.pb`.
### Test a model
We can check the quality of the trained model by running
@@ -261,12 +312,12 @@ Then we have three files
$ ls
conf.lmp graph-compress.pb in.lammps
-where conf.lmp gives the initial configuration of a gas phase methane MD simulation, and the file in.lammps is the lammps input script. One may check in.lammps and finds that it is a rather standard LAMMPS input file for a MD simulation, with only two exception lines:
+where `conf.lmp` gives the initial configuration of a gas phase methane MD simulation, and the file `in.lammps` is the lammps input script. One may check in.lammps and finds that it is a rather standard LAMMPS input file for a MD simulation, with only two exception lines:
pair_style graph-compress.pb
pair_coeff * *
-where the pair style deepmd is invoked and the model file graph-compress.pb is provided, which means the atomic interaction will be computed by the DP model that is stored in the file graph-compress.pb.
+where the pair style deepmd is invoked and the model file `graph-compress.pb` is provided, which means the atomic interaction will be computed by the DP model that is stored in the file graph-compress.pb.
One may execute lammps in the standard way
diff --git a/source/Tutorials/DeePMD-kit/learnDoc/run5minutes.md b/source/Tutorials/DeePMD-kit/learnDoc/run5minutes.md
index 8f17086..df1e7d2 100644
--- a/source/Tutorials/DeePMD-kit/learnDoc/run5minutes.md
+++ b/source/Tutorials/DeePMD-kit/learnDoc/run5minutes.md
@@ -18,13 +18,7 @@ What? Only three steps? Yes, it's that simple.
First, let's download and decompress the tutorial data:
```
- $ wget https://github.com/likefallwind/DPExample/raw/main/DeePMD-kit-FastLearn.tar
- $ tar xvf DeePMD-kit-FastLearn.tar
-```
-
-If you have trouble connecting github, you can download here:
-```
- $ wget https://gitee.com/likefallwind/dpexamples/raw/main/DeePMD-kit-FastLearn.tar
+ $ wget https://dp-public.oss-cn-beijing.aliyuncs.com/community/DeePMD-kit-FastLearn.tar
$ tar xvf DeePMD-kit-FastLearn.tar
```
diff --git a/source/conf.py b/source/conf.py
index 203e54a..31a9606 100644
--- a/source/conf.py
+++ b/source/conf.py
@@ -58,3 +58,5 @@
# relative to this directory. They are copied after the builtin static files,
# so a file named "default.css" will overwrite the builtin "default.css".
html_static_path = ['_static']
+
+latex_engine = 'xelatex'
steps.
The training parameters are given in the following
@@ -176,7 +208,8 @@ If everything works fine, you will see, on the screen, information printed every
DEEPMD INFO batch 10000 training time 6.41 s, testing time 0.01 s
DEEPMD INFO saved checkpoint model.ckpt
-They present the training and testing time counts. At the end of the 10000th batch, the model is saved in Tensorflow's checkpoint file model.ckpt. At the same time, the training and testing errors are presented in file lcurve.out.
+They present the training and testing time counts. At the end of the 10000th batch, the model is saved in Tensorflow's checkpoint file `model.ckpt`. At the same time, the training and testing errors are presented in file `lcurve.out`.
+The file contains 8 columns, form left to right, are the training step, the validation loss, training loss, root mean square (RMS) validation error of energy, RMS training error of energy, RMS validation error of force, RMS training error of force and the learning rate. The RMS error (RMSE) of the energy is normalized by number of atoms in the system.
$ head -n 2 lcurve.out
#step rmse_val rmse_trn rmse_e_val rmse_e_trn rmse_f_val rmse_f_trn lr
@@ -190,11 +223,29 @@ and
Volumes 4, 5 and 6, 7 present energy and force training and testing errors, respectively. It is demonstrated that after 140,000 steps of training, the energy testing error is less than 1 meV and the force testing error is around 120 meV/Å. It is also observed that the force testing error is systematically (but slightly) larger than the training error, which implies a slight over-fitting to the rather small dataset.
+One can visualize this file by a simple Python script:
+
+```py
+import numpy as np
+import matplotlib.pyplot as plt
+
+data = np.genfromtxt("lcurve.out", names=True)
+for name in data.dtype.names[1:-1]:
+ plt.plot(data['step'], data[name], label=name)
+plt.legend()
+plt.xlabel('Step')
+plt.ylabel('Loss')
+plt.xscale('symlog')
+plt.yscale('log')
+plt.grid()
+plt.show()
+```
+
When the training process is stopped abnormally, we can restart the training from the provided checkpoint by simply running
$ dp train --restart model.ckpt input.json
-In the lcurve.out, you can see the training and testing errors, like
+In the `lcurve.out`, you can see the training and testing errors, like
538000 3.12e-01 2.16e-01 6.84e-04 7.52e-04 1.38e-01 9.52e-02 4.1e-06
538000 3.12e-01 2.16e-01 6.84e-04 7.52e-04 1.38e-01 9.52e-02 4.1e-06
@@ -203,7 +254,7 @@ In the lcurve.out, you can see the training and testing errors, like
530000 2.89e-01 2.15e-01 6.36e-04 5.18e-04 1.25e-01 9.31e-02 4.4e-06
531000 3.46e-01 3.26e-01 4.62e-04 6.73e-04 1.49e-01 1.41e-01 4.4e-06
-Note that input.json needs to be consistent with the previous one.
+Note that `input.json` needs to be consistent with the previous one.
### Freeze and Compress a model
At the end of the training, the model parameters saved in TensorFlow's checkpoint file should be frozen as a model file that is usually ended with extension .pb. Simply execute
@@ -212,8 +263,8 @@ At the end of the training, the model parameters saved in TensorFlow's checkpoin
DEEPMD INFO Restoring parameters from ./model.ckpt-1000000
DEEPMD INFO 1264 ops in the final graph
-and it will output a model file named graph.pb in the current directory.
-The compressed DP model typically speed up DP-based calculations by an order of magnitude faster, and consume an order of magnitude less memory. The graph.pb can be compressed in the following way:
+and it will output a model file named `graph.pb` in the current directory.
+The compressed DP model typically speed up DP-based calculations by an order of magnitude faster, and consume an order of magnitude less memory. The `graph.pb` can be compressed in the following way:
$ dp compress -i graph.pb -o graph-compress.pb
DEEPMD INFO stage 1: compress the model
@@ -227,7 +278,7 @@ The compressed DP model typically speed up DP-based calculations by an order of
DEEPMD INFO Restoring parameters from model-compression/model.ckpt
DEEPMD INFO 840 ops in the final graph
-and it will output a model file named graph-compress.pb.
+and it will output a model file named `graph-compress.pb`.
### Test a model
We can check the quality of the trained model by running
@@ -261,12 +312,12 @@ Then we have three files
$ ls
conf.lmp graph-compress.pb in.lammps
-where conf.lmp gives the initial configuration of a gas phase methane MD simulation, and the file in.lammps is the lammps input script. One may check in.lammps and finds that it is a rather standard LAMMPS input file for a MD simulation, with only two exception lines:
+where `conf.lmp` gives the initial configuration of a gas phase methane MD simulation, and the file `in.lammps` is the lammps input script. One may check in.lammps and finds that it is a rather standard LAMMPS input file for a MD simulation, with only two exception lines:
pair_style graph-compress.pb
pair_coeff * *
-where the pair style deepmd is invoked and the model file graph-compress.pb is provided, which means the atomic interaction will be computed by the DP model that is stored in the file graph-compress.pb.
+where the pair style deepmd is invoked and the model file `graph-compress.pb` is provided, which means the atomic interaction will be computed by the DP model that is stored in the file graph-compress.pb.
One may execute lammps in the standard way
diff --git a/source/Tutorials/DeePMD-kit/learnDoc/run5minutes.md b/source/Tutorials/DeePMD-kit/learnDoc/run5minutes.md
index 8f17086..df1e7d2 100644
--- a/source/Tutorials/DeePMD-kit/learnDoc/run5minutes.md
+++ b/source/Tutorials/DeePMD-kit/learnDoc/run5minutes.md
@@ -18,13 +18,7 @@ What? Only three steps? Yes, it's that simple.
First, let's download and decompress the tutorial data:
```
- $ wget https://github.com/likefallwind/DPExample/raw/main/DeePMD-kit-FastLearn.tar
- $ tar xvf DeePMD-kit-FastLearn.tar
-```
-
-If you have trouble connecting github, you can download here:
-```
- $ wget https://gitee.com/likefallwind/dpexamples/raw/main/DeePMD-kit-FastLearn.tar
+ $ wget https://dp-public.oss-cn-beijing.aliyuncs.com/community/DeePMD-kit-FastLearn.tar
$ tar xvf DeePMD-kit-FastLearn.tar
```
diff --git a/source/conf.py b/source/conf.py
index 203e54a..31a9606 100644
--- a/source/conf.py
+++ b/source/conf.py
@@ -58,3 +58,5 @@
# relative to this directory. They are copied after the builtin static files,
# so a file named "default.css" will overwrite the builtin "default.css".
html_static_path = ['_static']
+
+latex_engine = 'xelatex'