-
Notifications
You must be signed in to change notification settings - Fork 3
Open
Description
Hi,
I have some results I cannot really explain when looking at the raw pvalue distribution.
Here is my framework from the csaw book ;
param <- readParam(restrict=paste0("chr", 1:22), minq=20)
win.data <- windowCounts(bams, width=2000, spacing=1000, ext=125, param=param)
bins <- windowCounts(bams, bin=TRUE, width=10000, param=param)
## Local filtering
neighbor <- suppressWarnings(resize(rowRanges(win.data), width=5000, fix="center"))
wider <- regionCounts(bams, regions=neighbor, ext=frag.len, param=param)
filter.stat <- filterWindowsLocal(win.data, wider)
hist(filter.stat$filter, main="", breaks=50,
xlab="Background abundance (log2-CPM)")
abline(v=log2(2), col="red")
keep <- filter.stat$filter > log2(2)
summary(keep)
filtered.data <- win.data[keep,]
## Normalize on bg
filtered.data <- normFactors(bins, se.out=filtered.data)
## Differential analysis
y <- asDGEList(filtered.data)
design <- model.matrix(~ 0 + splan.sub$GROUP)
colnames(design) <- c("EGF", "untreated")
y <- estimateDisp(y, design)
fit <- glmQLFit(y, design, robust=TRUE)
contrast <- makeContrasts(EGF-untreated, levels=design)
res.csaw <- glmQLFTest(fit, contrast=contrast[,1])
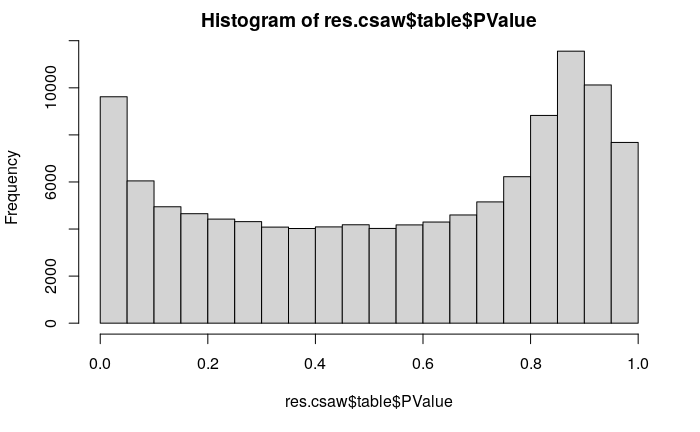
hist(res.csaw$table$PValue)
And here is my raw pvalues distribution :
As I do see this peak for extreme pvalues, I decided to be more stringent on the filtering. Usually, from my experience on RNA-seq data, it should help solving this issue.
However, I do have exactly the opposite effect !
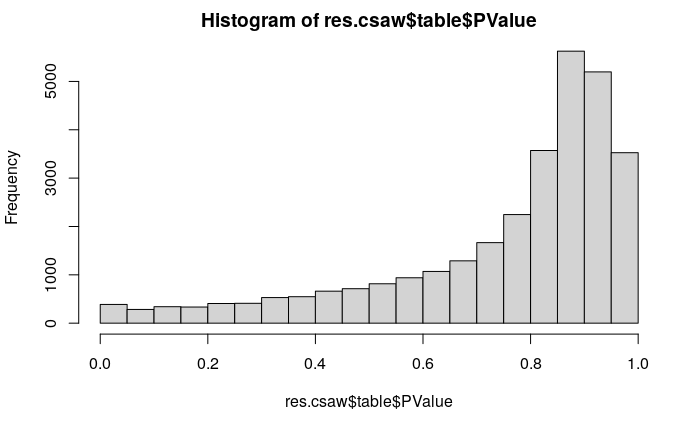
keep <- filter.stat$filter > log2(5)
summary(keep)
filtered.data <- win.data[keep,]
I was wondering how we could explain this behaviour ?
I have the same effect using global/local filtering.
Many thanks
Reactions are currently unavailable
Metadata
Metadata
Assignees
Labels
No labels